Sonia Patel posted this recently AI in Healthcare: Hype or Reality? | Sonia Patel FBCS posted on the topic | LinkedIn
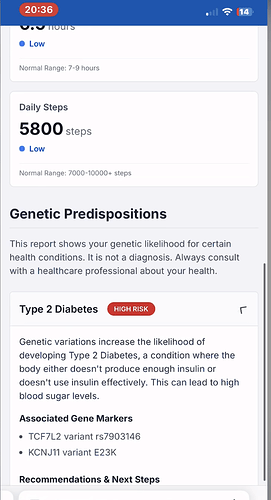
I was intrigued by the use of myDNA DNA Reporting Services I myDNA Precision Health together with NHS App, it showed genes (Variants) and the possible conditions this implies (in standards this part is called Diagnostic Implication, the screen shot calls this Genetic Predispositions). I really liked how this post presented the problems and tech.
Then a few days later my daughter mentioned she had FOXO3 (Variant) and this came from both parents (Homozygous), which also has implications.
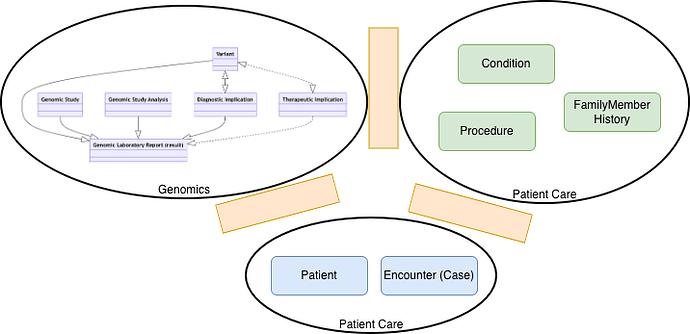
Since then I’ve started to look at how this would be represented in a Genomic system North West Genomics Test Report - NHS North West Genomics v0.0.8 ← this gets a bit complicated but I’m aiming to focus on how this was presented in the linkedin post and how my daughter described it.
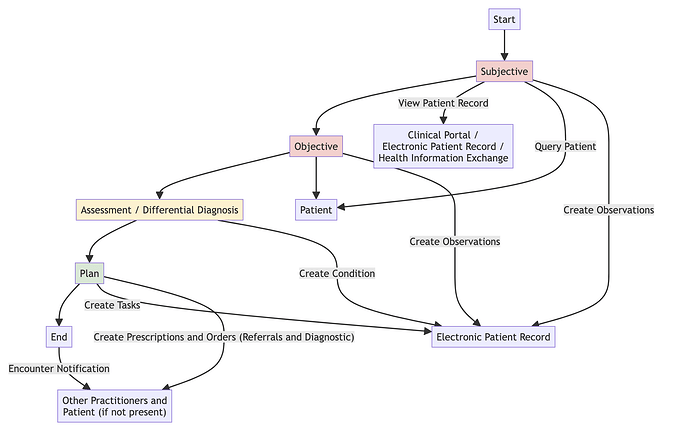
Where I’m up to at the moment is wondering how a GP or Consultant would record this in an EPR system. (I know how EPIC would share this thanks to it’s open api documentation Specifications - Epic on FHIR ) and how to get it into an EPR (from US supplier) using HL7 v2 ORU_R01)
I think a GP would either record a clinical entry (Observation) or problem (Condition), for Observation I think the codes are:
Observation
- Genomic Finding Detected - NHS North West Genomics v0.0.8
- Genomic Finding - NHS North West Genomics v0.0.8
- Genomic Disorder Carrier - NHS North West Genomics v0.0.8
Probably need access to a clinical coder in primary or secondary care but I’m currently in diagnostic testing. Probably making this difficult as I’m employed as a developer and need to pass this up the chain.
Any views?